Overview
The focus of the iReceptor project is to federate large Adaptive Immune Receptor Repertoire (AIRR-seq) data repositories from multiple laboratories and enable researchers to easily and efficiently perform complex analyses on these federated repositories. One of the key design components of iReceptor is its distributed data model. We call this the AIRR Data Commons. A distributed data model, although difficult to support, is we believe critical to the success of research in this area. This is for two main reasons:
- Next generation sequencing has caused an explosion in the data available to labs that are carrying out immunogenetics research. In order to answer complex immunogenetic questions, these labs need to collaborate in a variety of ways. Although large scale repositories for AIRR-seq data exist, it is our belief that it is not practical to provide a single repository (or even a small number of large repositories) at the scale that will be effective in enabling the types of collaboration that need to occur. A distributed data model means that each lab needs to store a relatively small amount of data. A Scientific Gateway that enables the federation of such data sets means that complex research questions (through queries across distributed AIRR-seq data sets) can be answered.
- AIRR-seq data is invariably patient data, and therefore needs to be treated with confidentiality and security. Data use typically goes through institutional ethics boards and the data stewards at given institutions need to be confident that the data is treated securely. A distributed data model enables the data steward to store, monitor, and share data while at the same time having explicit and direct control over who has access to that data.
The goal of the iReceptor project is to hide the technical complexities of the above problem, while at the same time empowering immunogenetics researchers to perform very sophisticated (and in many cases, computationally expensive) analyses on federated data from multiple, distributed AIRR-seq repositories.
Architectural components
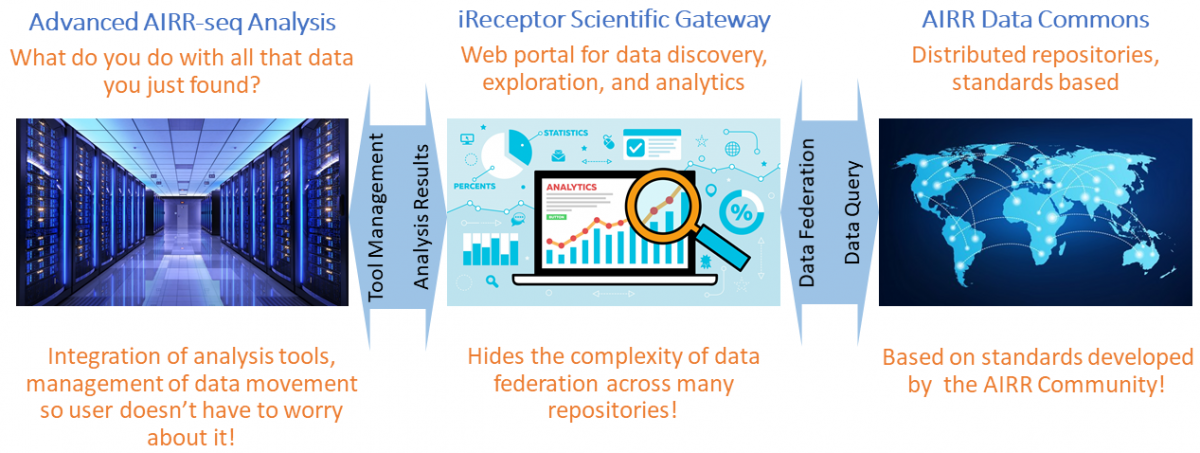
There are a number of architectural components of the iReceptor Platform:
- An iReceptor AIRR-seq data model and repository design that builds on the AIRR Community's Minimal Standard for AIRR-seq data (MiAIRR). The MiAIRR standard encompasses the basic sequence data, meta-data about the source of the sequence data (subject, lab, experiment, and sample data), and annotation data from annotation tools such as IMGT's vQuest, igblast, and MiXCR. This data model has been developed by the AIRR Community and implemented by the iReceptor team.
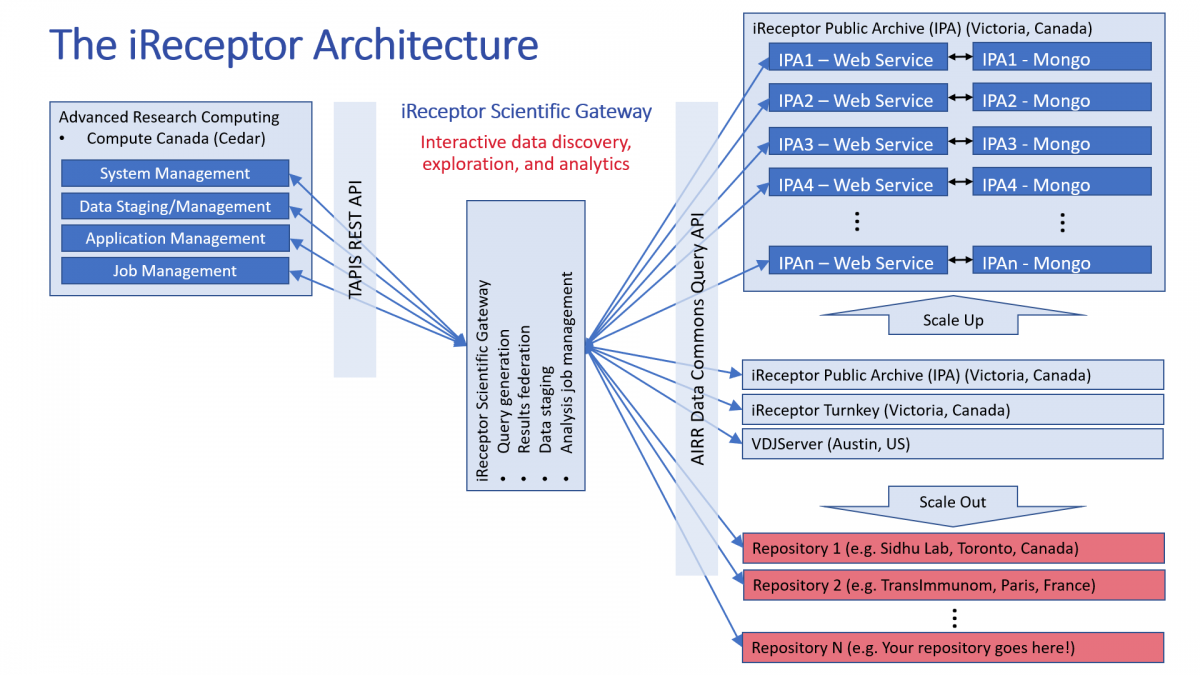
- A data repository service that provides access to AIRR-seq data repositories that use the MiAIRR data standard. This service will allow research labs that have an AIRR-seq repository that supports the MiAIRR data model to expose parts of their repository to external clients. Access to this data is provided through the implementation of the AIRR Data Common API, a web-based application programming interface that allows queries to be made against an iReceptor repository. Through this service, the iReceptor Gateway will enable researchers to pose queries across multiple, distributed, AIRR-seq repositories and to federate the results of those queries for analysis.
- An AIRR Data Commons. The above two components, in combination, provide the ability to create a network of international AIRR compliant data repositories that we call the AIRR Data Commons.
- A scientific gateway web platform that can federate distributed AIRR-seq repositories and perform complex analyses on this federated data. The iReceptor Gateway will not only link distributed AIRR-seq repositories but it will also enable researchers to perform complex analyses on high end computational systems (for example, systems in Compute Canada). Note, a key component of the iReceptor Gateway is that it hides the complexity of the repository queries, data staging of the federated data, and the advanced computation from the end user.
- A set of analysis services that enable AIRR-seq researchers to perform computationally expensive analyses on advanced computational infrastructure. Using the AGAVE/Tapis Science-as-a-Service architecture, the iReceptor Gateway enables extensible and flexible data movement and advanced computation on a wide range of HPC platforms including Compute Canada infrastructure.
The iReceptor project combines all of these components into the following resources that the general research community might be interested in:
- The iReceptor Turnkey is software that researchers can download to help manage their own data in their own AIRR compliant repository.
- The iReceptor Scientific Gateway is a web based user interface that allows researchers to search and explore the entire AIRR Data Commons.
A more detailed diagram of the iReceptor architecture is given below. In this diagram, there are a set of distributed AIRR-seq repositories (the AIRR Data Commons), existing at collaborating international institutions (e.g Simon Fraser University, the University of Texas (UT) Southwestern Medical Center, and Sorbonne University). Each centre maintains its own repository and has explicit control over that labs data. Through the us of the AIRR Data Commons API, external users can query these repositories. Using the iReceptor Gateway, researchers can pose complex immunogenetics research questions across the repositories in the AIRR Data Commons. The iReceptor Gateway federates the results of the queries across the distributed repositories and stores that data in a researcher's workspace. That data can then be staged to a large HPC resource for computation. After the computation is completed, the iReceptor Gateway stages the analysis results back to the researcher's workspace. Using this model, a researcher can then perform iterative data exploration and analysis across all of their collaborators data.