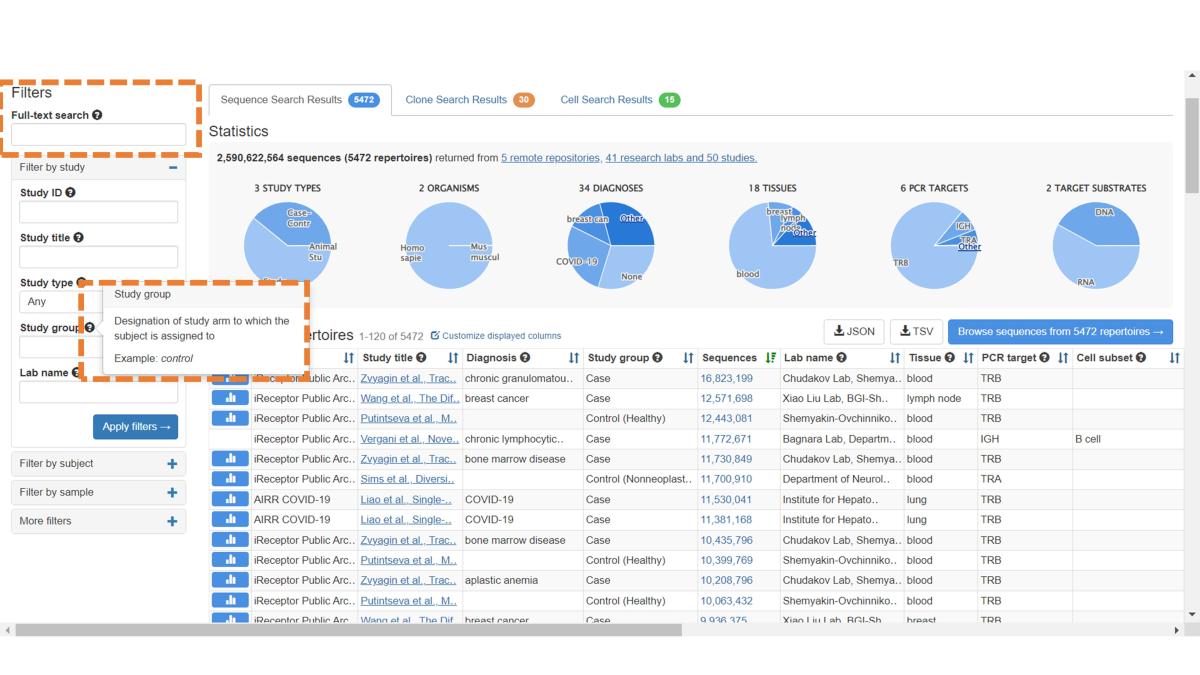
By default, the Repertoire Metadata Search lands on the Sequence Search Results tab, indicated by blue summary graphs in the Statistics panel. All available metadata are presented, but you can filter based on subject/study/sample fields via boxes on the left-hand side of the search page. For now, filters with free-text entries perform a substring match (no regular expressions). For more information on filters, hover over the black question marks.
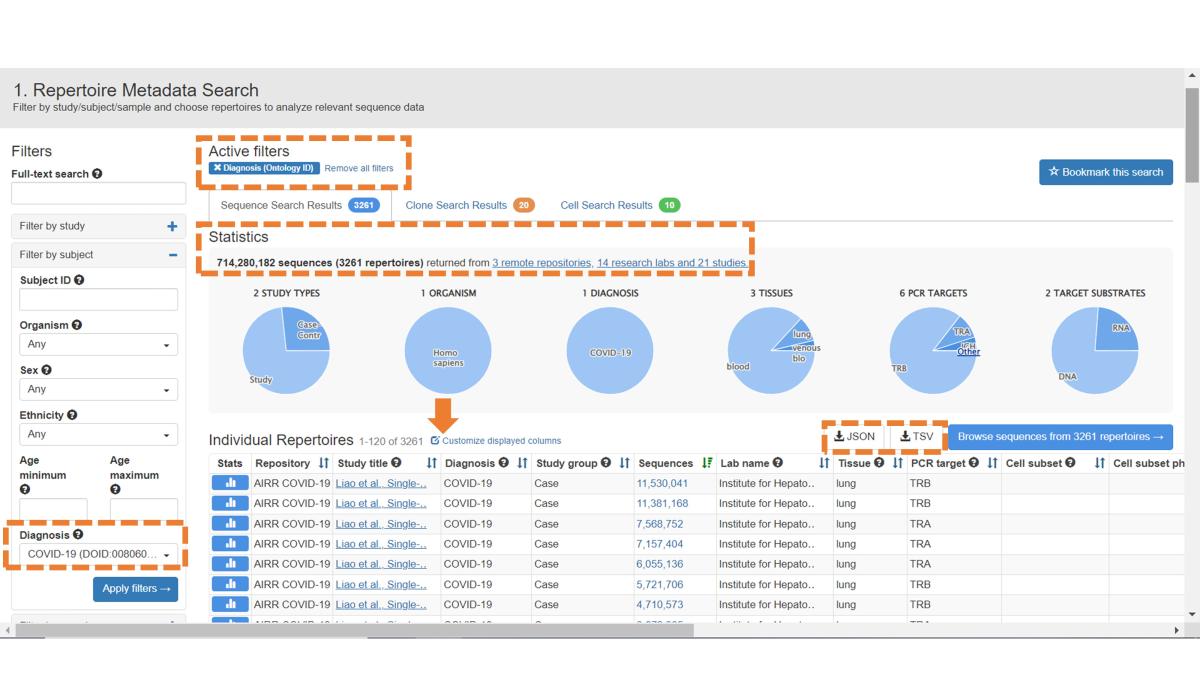
In this example, we wish to explore sequence data from COVID-19 subjects, so we’ve applied the Diagnosis filter (ontology based). The updated results are shown. The Active Filters panel now displays the engaged filter, and Statistics have been updated to reflect the filtered data. The default metadata fields are displayed (scroll all the way to the right), and you can download this metadata by clicking on the JSON or TSV button, depending on your desired format.
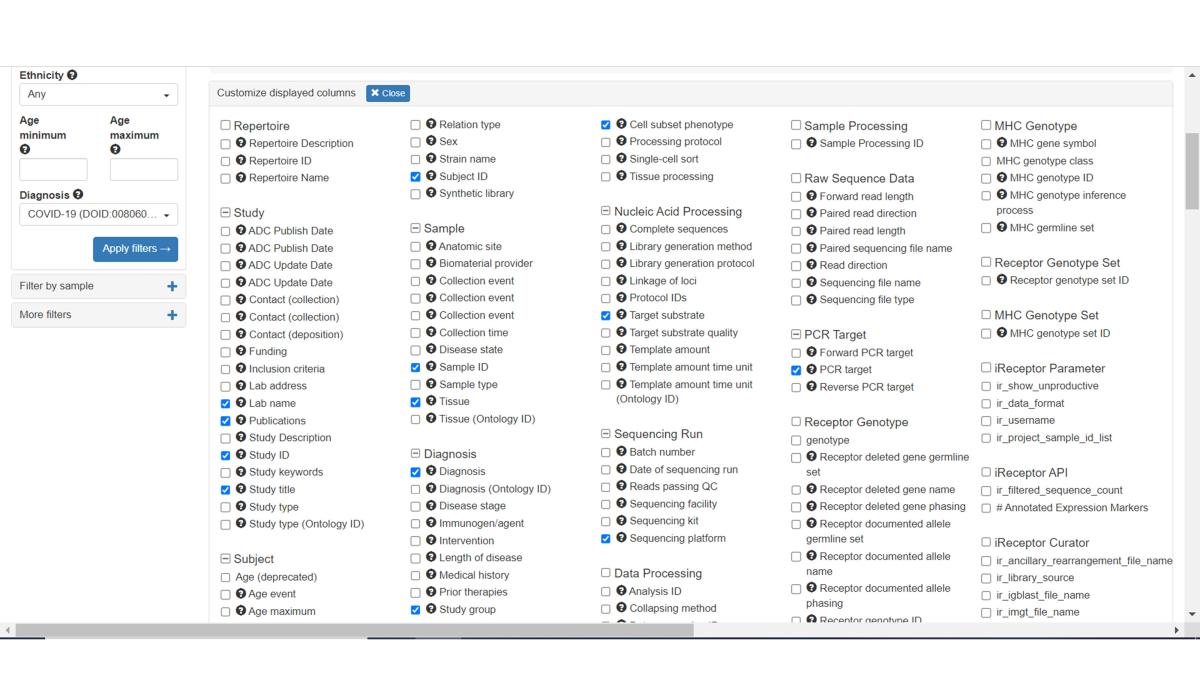
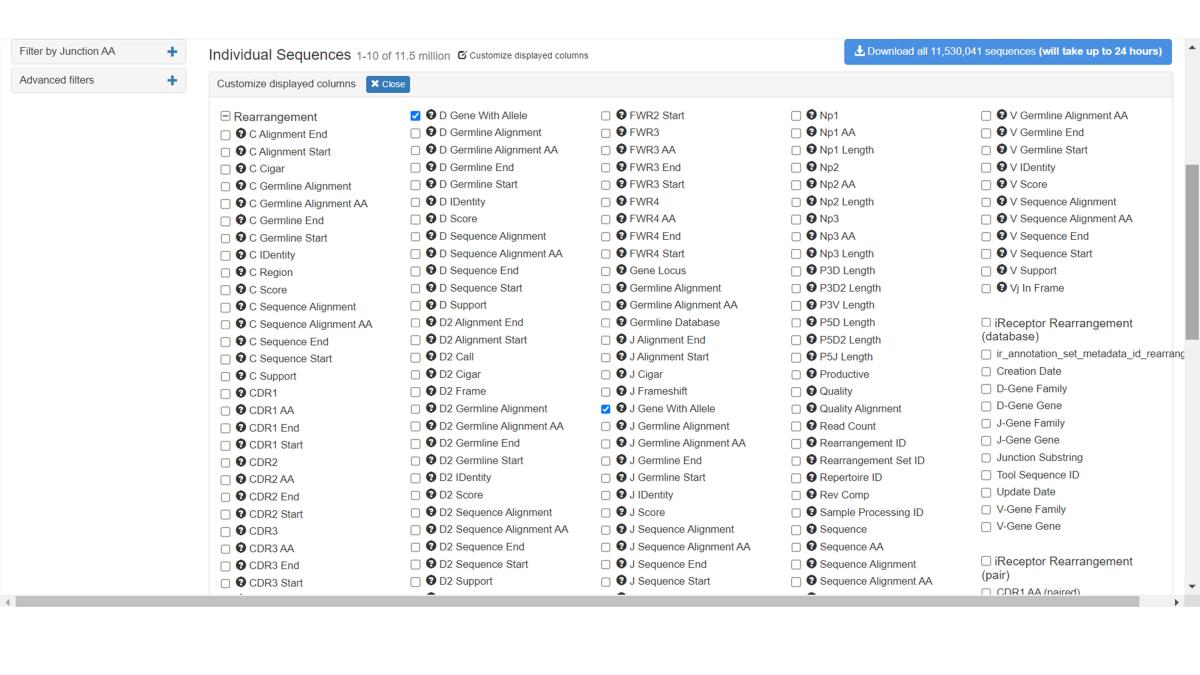
Create a customized view including any of the 80+ MiAIRR fields by clicking on the Customize Displayed Columns button. Newly added columns will be added to the right-most side of the table.
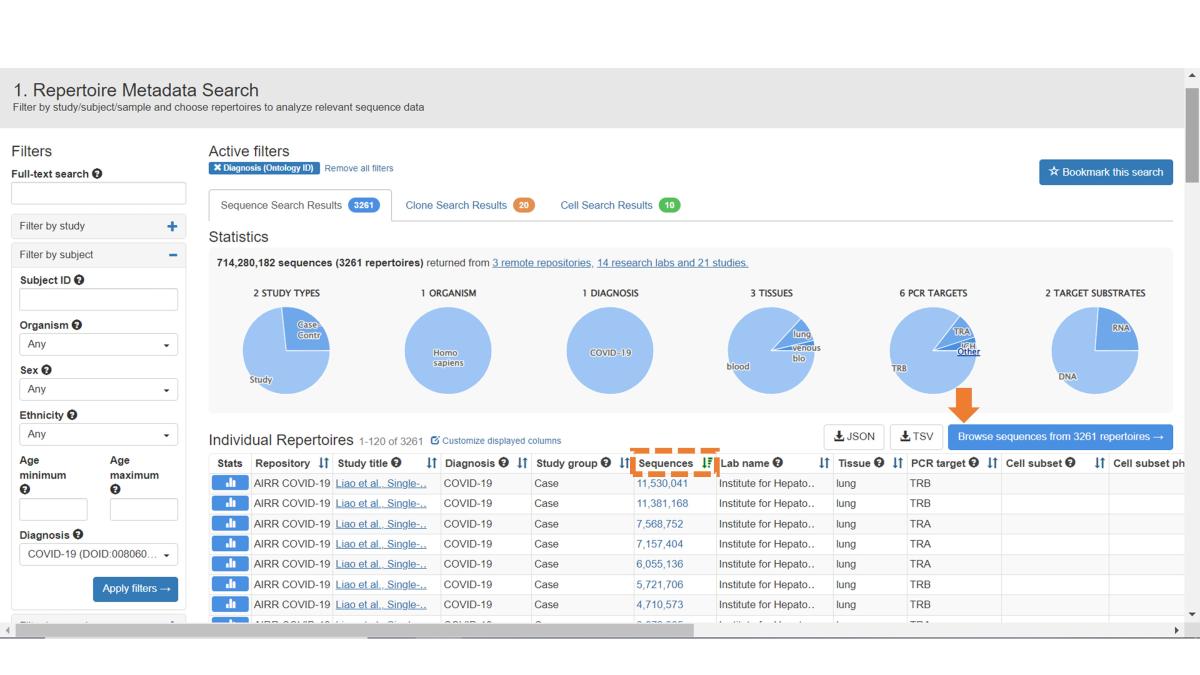
You can browse sequences from individual samples by clicking on the respective sample in the Sequences field. Samples are listed in descending order based on sequence count, but you can customize this by clicking on the name of your desired sort field. To browse sequences of all samples in the filtered list, click on the Browse Sequences From All Repertoires button.
In this example, we will browse sequences from the first sample in the list. This opens a new Sequence Search page, and the navigation menu indicates we have entered stage 2 of the search workflow. You can still go back to the metadata page and see/remove any filters applied there.
The first 10 individual sequences from your search results are shown. In the left-hand panel, you can filter sequences based on V, D, and J genes, CDR3 sequence and length, and remove unproductive rearrangements.
Click on the Download button to obtain all sequences in AIRR format, as well as an info file detailing search criteria and location of sequences. Note that downloads of over 500M sequences are not currently available.
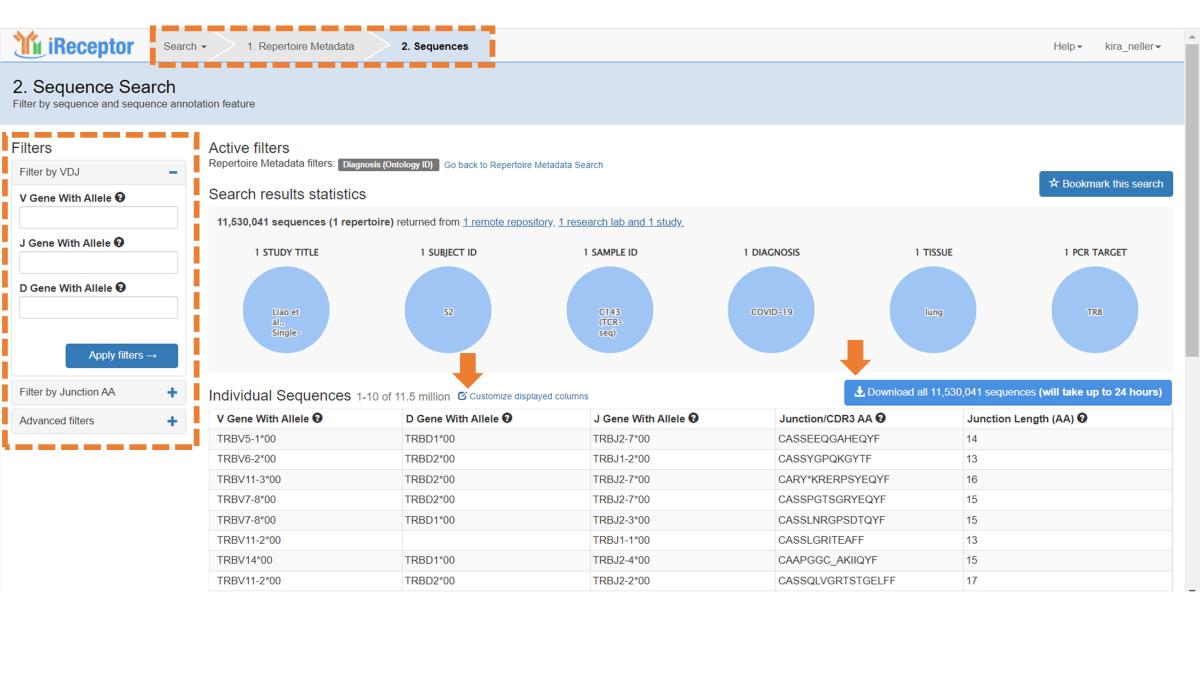
As with metadata, you can customize the displayed sequence annotation fields.
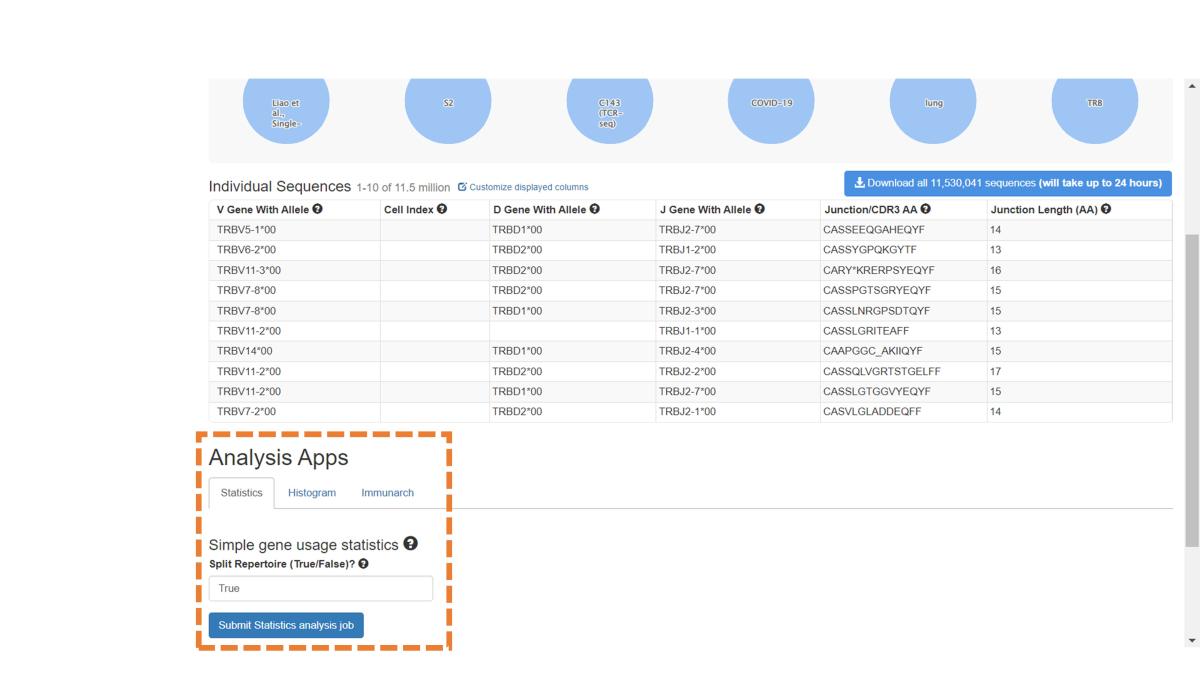
Scroll down to the bottom of the Sequences page to see the Analysis Apps available for sequence data. The Statistics App provides an overview of gene usage and CDR3 length, the Histogram App can be run on any chosen AIRR field, and Immunarch produces a range of summary plots, including diversity analyses. Clicking on the Submit button for any App will submit an analysis job. See the Analysis Page for more information.
- Log in to post comments
