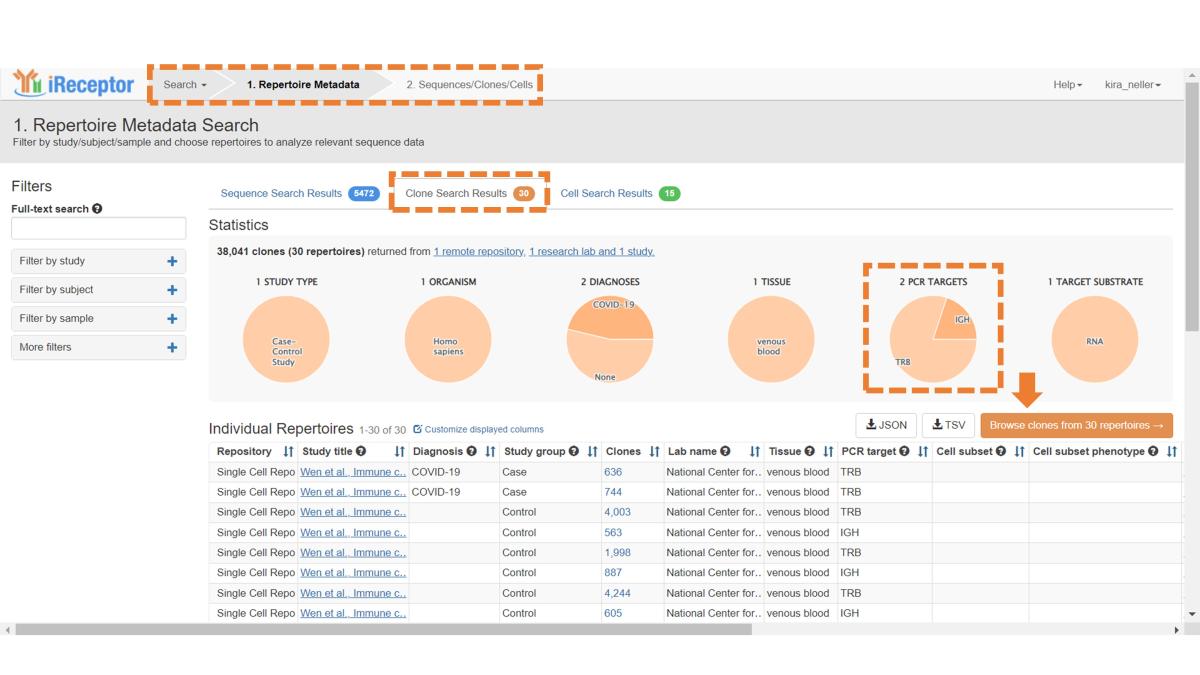
From the Repertoire Metadata Search page (which can be accessed via the top navigation menu if you just completed the Sequence workflow), click on the Clone Search Results tab. The updated orange colour scheme confirms you have accessed the clone workflow and are viewing metadata for clone samples. In this context, “clone” refers to a group of BCR or TCR rearrangements arising from a single ancestral B or T cell, inferred from clustering and assignment algorithms.
As shown in the Statistics panel, there are only two loci available: IGH and TRB. This is because the example data comes from a 10x single-cell study, and though paired chain information is available, the AIRR Standard cannot represent clones with multiple chains as of yet – we hope to provide this ability soon. For more information on creating an AIRR Clone file from 10x VDJ data, see here. Note that users submitting non-paired data can do so for any locus (IGH/IGK/IGL/TRA/TRB/TRD/TRG), as the above pertains only to representation of clones with paired chains.
As with Sequence-level metadata, from this page you can apply filters, customize the displayed metadata, and download metadata. Click the Browse Clones button to enter stage 2 of the search.
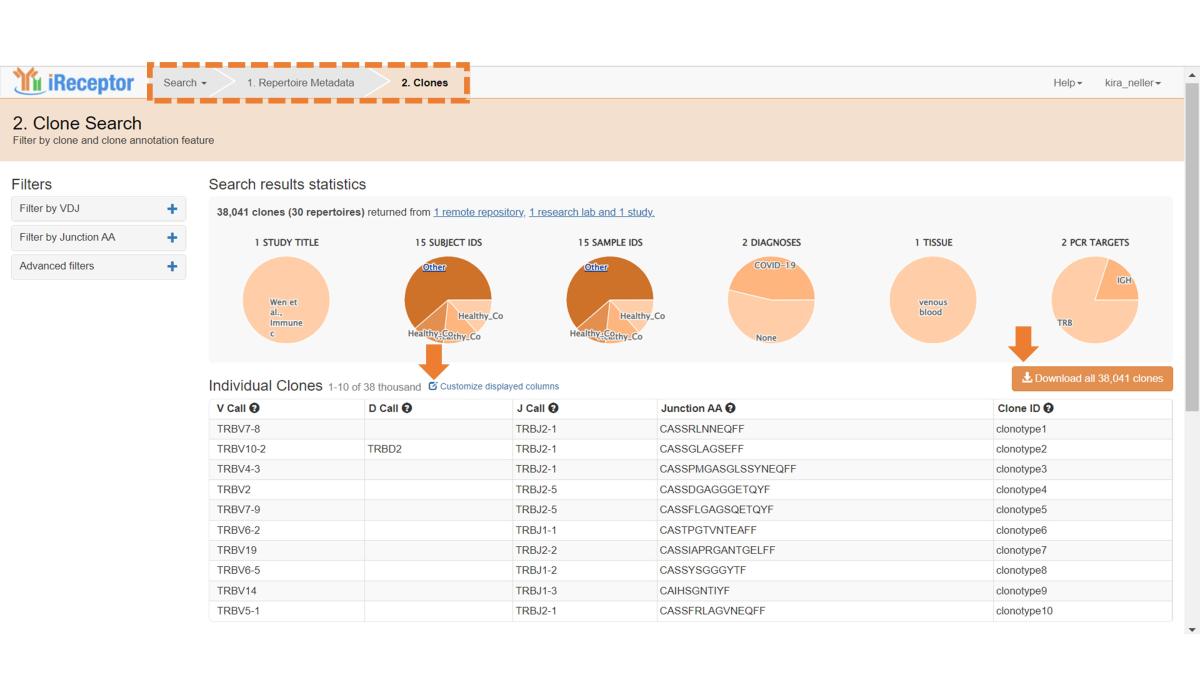
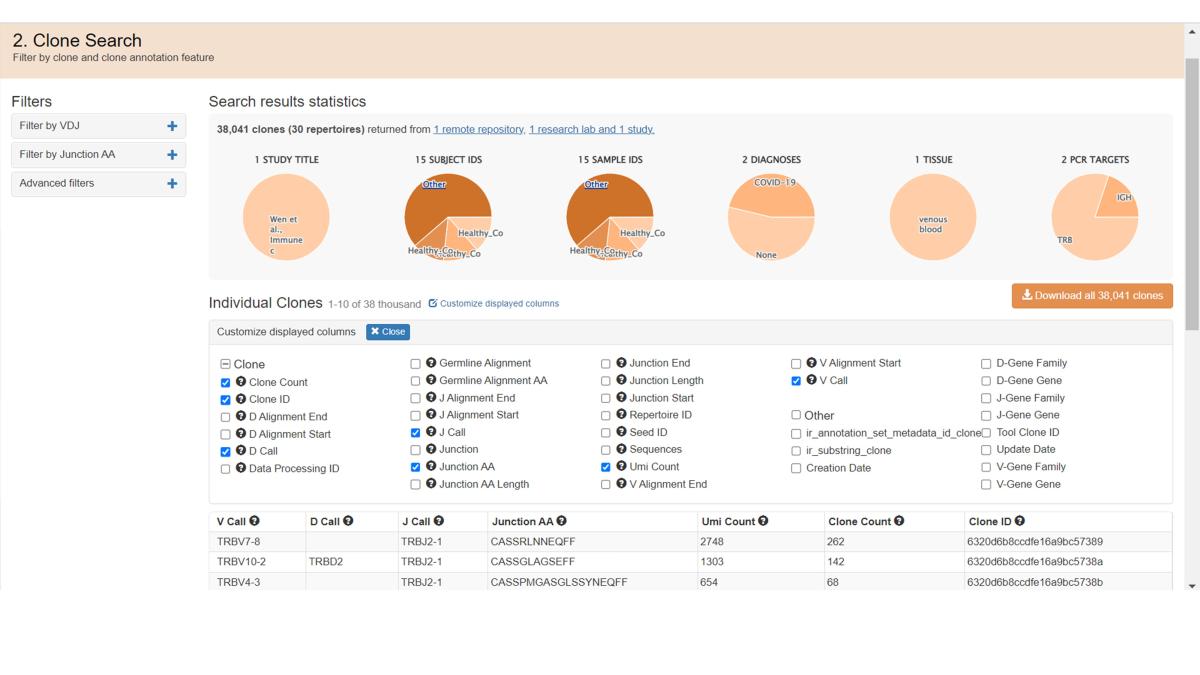
This opens a new Clone Search page, analogous to the Sequence Search page, and provides the same features as described above – you can filter clone sequences, customize displayed annotation fields, and download annotated clone data).
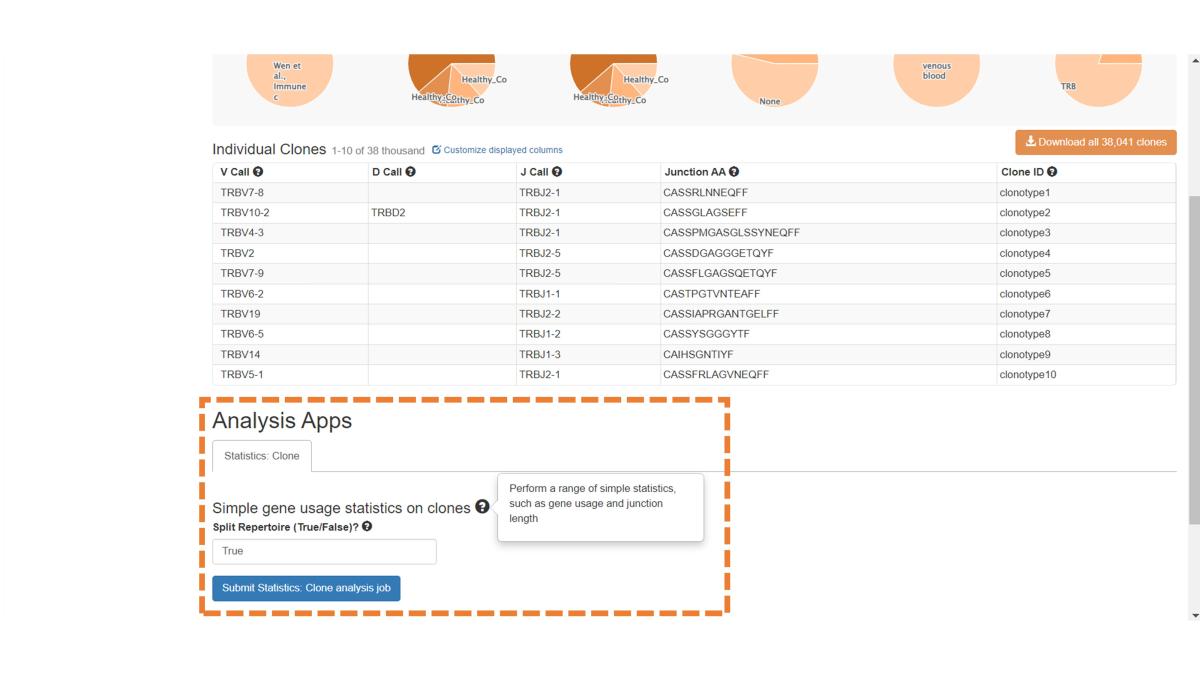
Scroll down to the bottom of the Clones page to see the available Analysis Apps, which provide gene usage statistics for clones. See the Analysis Page for more information on running jobs.
- Log in to post comments
